Carl Bauer's group uses the cryoTEM facility to visualize photoreceptor proteins that are involved in bacterial vision. The laboratory uses a combination of biochemistry, X-ray crystallography and now cryo-electron microscopy to decipher interactions that occur between three proteins involved in bacterial vision.
Some Current Research Using Electron Microscopy Center Facilities
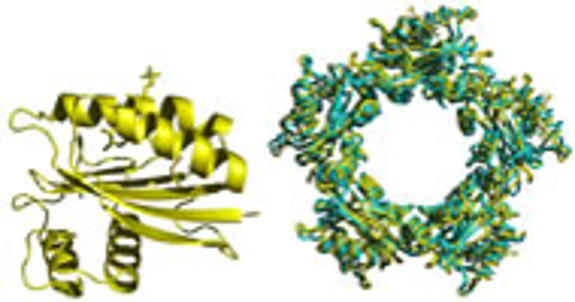
Stephen Bell's group studies various aspects of the molecular biology of Archaea. They are interested in the structural organization of the cell itself and the mechanisms of cell division. In addition, the laboratory is starting to use cryoTEM to elucidate the architecture of the macromolecular complexes that mediate gene transcription and DNA replication.
Lyudmila Bronstein's group focuses on the formation of inorganic nanoparticles (NPs), their structure, functionalization and applications including catalysis and biorelated studies. Using HRTEM, we will determine the structure of inorganic crystals. Elemental maps of compound NPs (quantum dots, bimetallic NPs, etc.) and their functional shells will be obtained using STEM/EDX capabilities.
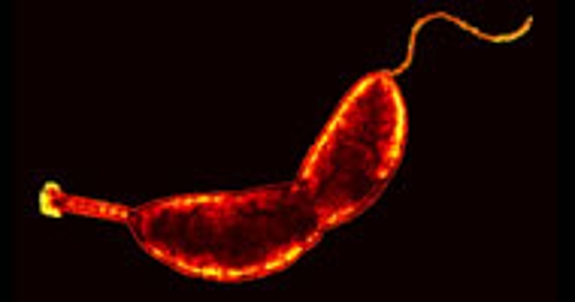
Bacteria can adopt a myriad of shapes and the mechanisms that generate those changes are only beginning to be understood. Yves Brun's group uses electron cryo tomograpghy (cryoET) to study the process by which Caulobacter crescentus and related bacteria synthesize a thin extension of the cell envelope called the stalk or prostheca. This process provides a tractable example of a specific morphological change.
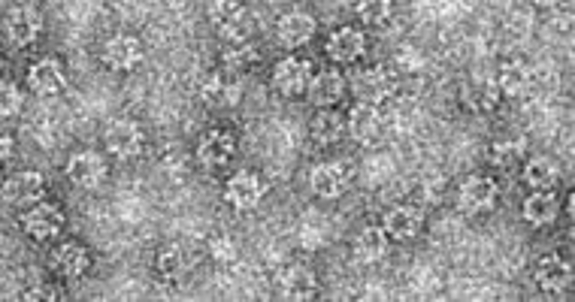
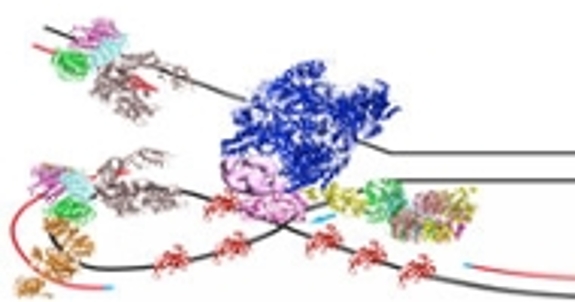
Lingling Chen's group studies protein-protein and protein-DNA interactions in biological processes. CryoEM is useful in providing the overall architecture of the macromolecular arrangements.
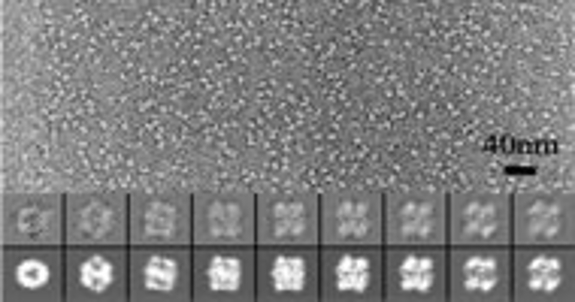
Cheng Kao's group uses the cryoEM facility to analyze the architecture of viruses and perform single particle reconstruction of protein and protein-RNA complexes.

Bacteria swim by rotating helical, propeller-like, flagella. Flagellar structure is complex and poorly understood in the Gram positive bacterium Bacillus subtilis. Daniel Kearns' group will genetically dissect components of flagellar assembly and use cryoEM to determine the structural consequences of flagellar mutants. They are especially interested in determining the structure of the flagella motor.
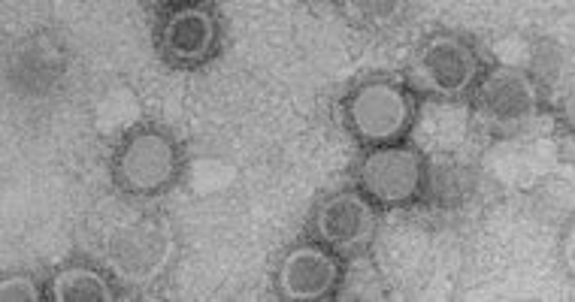
Tuli Mukhopadhyay's group uses cryoEM to determine the structure of different membrane bound RNA viruses (e.g., Sindbis virus). It is possible to learn how the virus carries out different functions using these structures.

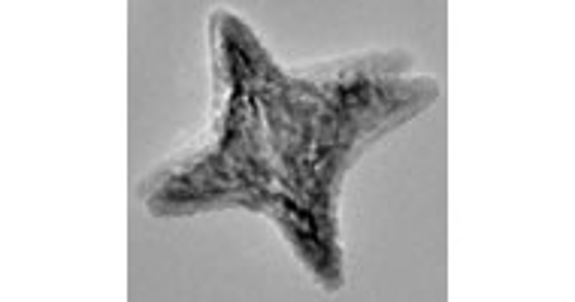
Sara Skrabalak's group focuses on the synthesis and application of architecturally and compositionally complex inorganic solids. The EM Center is instrumental in characterizing the dispersion of different components by elemental mapping using STEM/EDX.
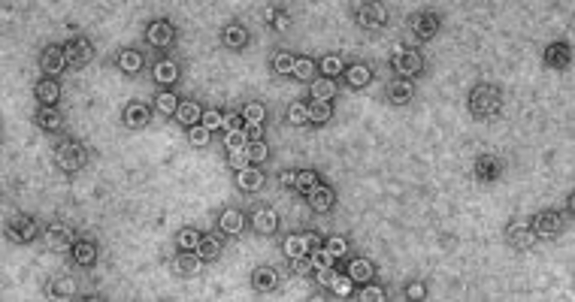
Adam Zlotnick's group takes viruses apart and puts them back together. The self-assembly of virus capsid proteins can be related to virus biology, development of anti-viral small molecules, and production of virus-based nanostructures. We will use cryoEM to examine the molecular details of naturally occurring and synthetic virus capsid protein structures.

