The deep staining procedure described below prevents (or significantly limits) the collapse seen when many purified biological specimens are negatively stained and air-dried using conventional techniques.
Deep Negative Stain
The following materials are used:
- Ammonium molybdate ( (NH4)2MoO4 )
- Trehalose (aka, α-D-Glucopyranosyl β-D-glucopyranoside)
Prepare the following solutions from these materials:
- 16% (w/v) Ammonium molybdate, pH 7.0
- place 0.16 g ammonium molybdate in a 1.5 ml Eppendorf tube
- add 0.8 ml distilled water and dissolve all the ammonium molybdate
- add a few drops of 10 N NaOH and adjust the pH to 7.0 (using pH paper)
- bring the final volume to 1.0 ml filter this solution through a 0.2 micron
- filter using a syringe
- 4% (w/v) Ammonium molybdate in 1% (w/v) trehalose, pH 7.0
- place 0.04 g ammonium molybdate in a 1.5 ml Eppendorf tube
- add 0.01 g trehalose
- add 0.8 ml distilled water and dissolve all the ammonium molybdate
- add a few drops of 10 N NaOH and adjust the pH to 7.0 (using pH paper)
- bring the final volume to 1.0 ml
- filter this solution through a 0.2 micron filter using a syringe
Always prepare these solutions immediately before use!
Use the following staining procedure:
- place an ~50 μl drop of either stain solution onto a piece of Parafilm
- apply 3 μl of sample to a glow-discharged grid
NOTE: holey, lacy and continuous carbon grids all work well, and the holey and lacy carbon support films probably eliminate some deformation due to interactions of the sample with the support film. - wait ~25 s (this will depend on the sample and the type of grid)
- blot (using filter paper) from the edge of the grid.
- immediately place the grid onto the stain droplet on the Parafilm sheet with the sample side facing the staining solution
- wait 10 s and blot again using filter paper
- at this point, there should be no stain visible on the grid surface
- if the surface does still appear wet, gently wave the grid in the air to speed up evaporation
Samples prepared this way should be examined as soon as possible (immediately if that option is available) in the electron microscope.
The details of this procedure and the deep-stain-embedded images shown below were kindly supplied by Che-Yen (Joe) Wang, and people should contact him for additional details and help with this procedure.
The first row of images show empty hepatitis B virus (HBV) capsids embedded in ammonium molybdate and trehalose that spans a 2 μm hole in a Quantifoil grid. The first and third images show a bit of the carbon support film in one corner of the image, and highlight the fact that these deep stain embedded virus capsids look much better when they do not sit on the support film.
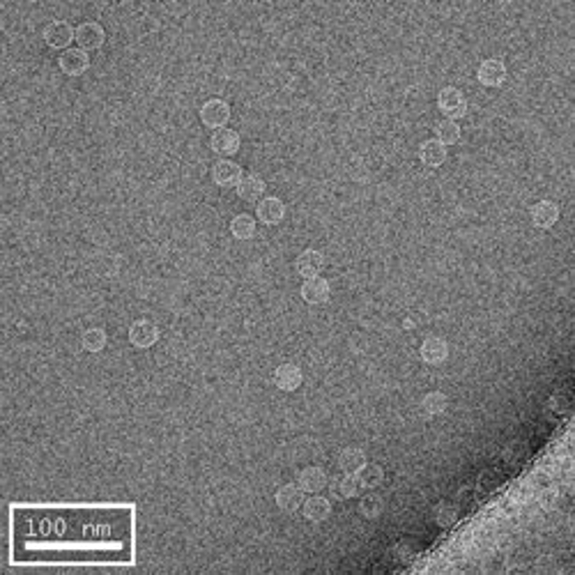
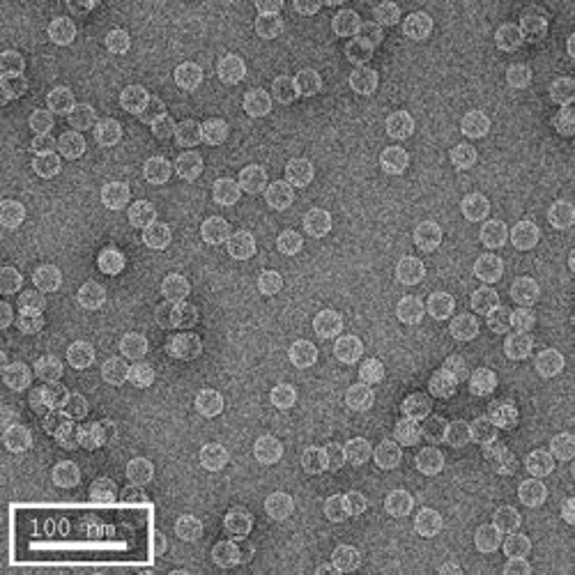
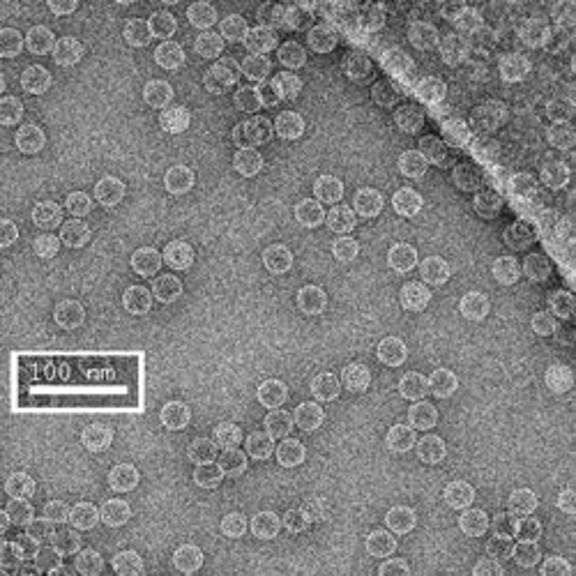
Compare the images above with those shown below. This second set of images shows similar HBV capsids that were conventionally stained on a continuous carbon support film using either uranyl acetate (upper row) or methylamine vanadate (NanoVan) (lower row). When conventionally stained images are compared to deep stain embedded images at the same defocus, the deep stain embedded images appear lower contrast (and are more similar to frozen, hydratred images). This is likely to be a result of the thickness of the stain layers. There is significantly less variability in images of deep stain embedded HBV capsids, especially at the edge of the capsids and across the fields of stain.
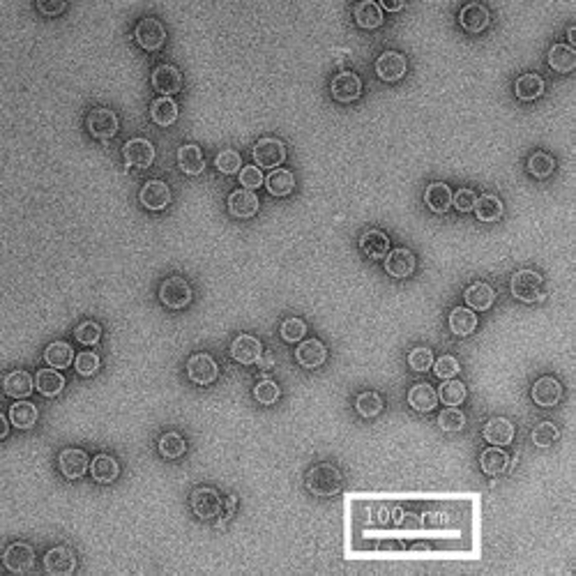
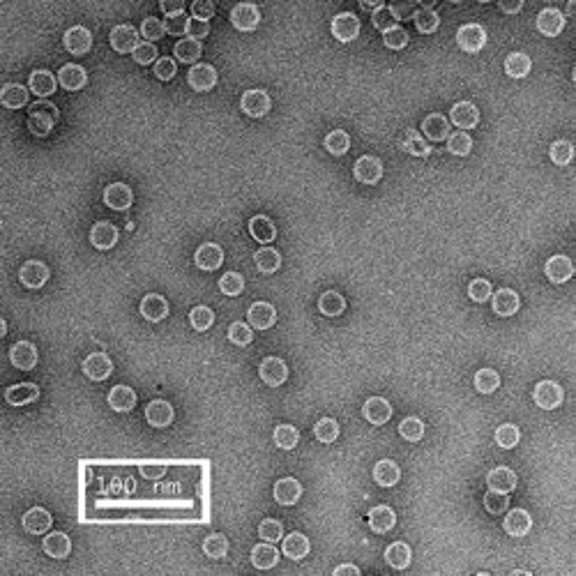
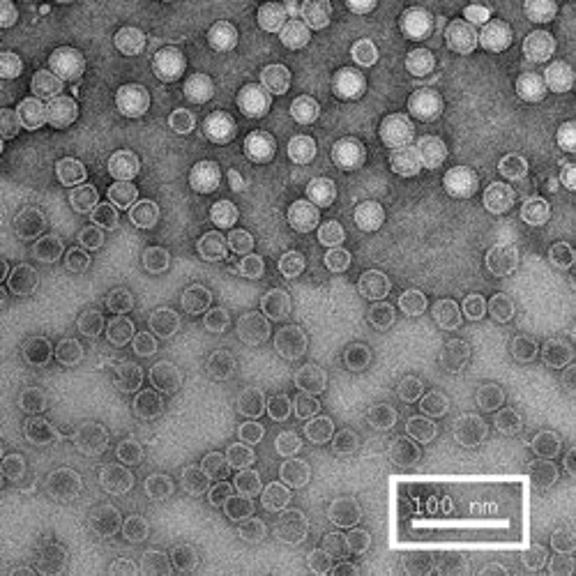
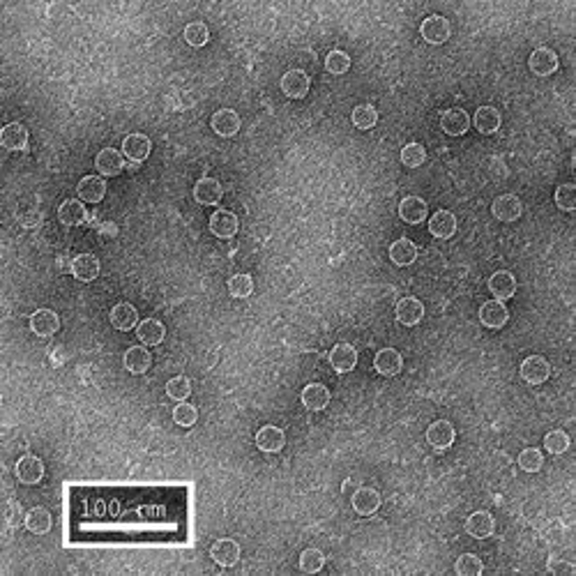
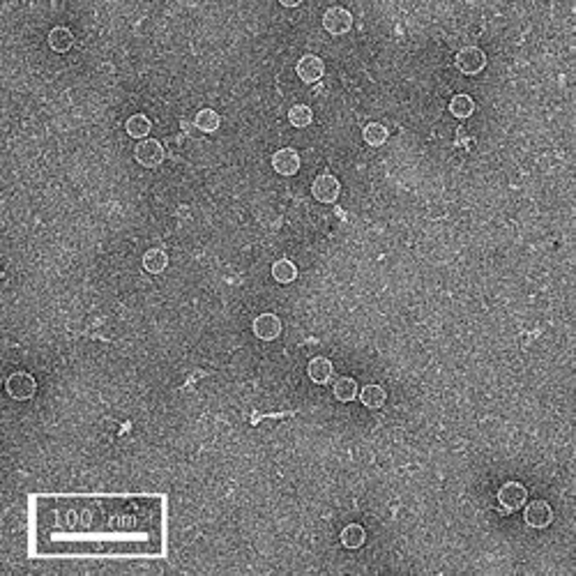
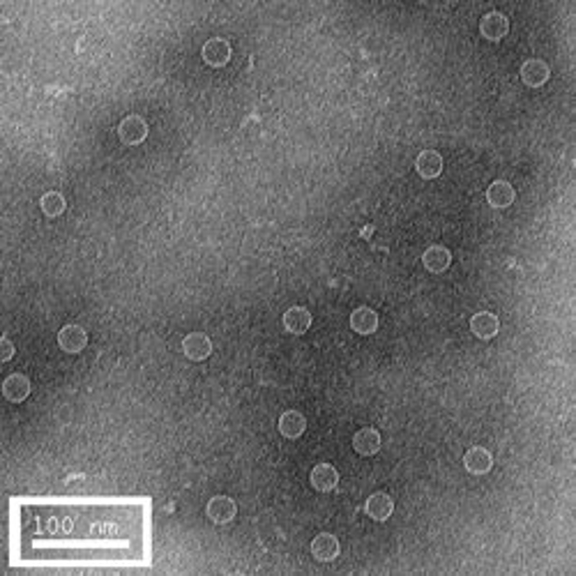
When negatively stained samples of spherical viruses such as HBV are tilted in the electron microscope, it is clear that there is a significant amount of collapse. The use of deep staining procedures limits this collapse to some extent, but does not completely prevent it.

